Cells control the expression levels of their proteins primarily by transcriptional regulation, a process where special signaling proteins called transcription factors (TFs), find and bind regulatory sequences on the DNA, thereby activating or repressing target genes. Transcription factors also regulate each other and respond to external signals, forming a regulatory network that orchestrates the cellular response to environmental cues.
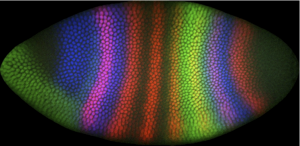
Image of an early Drosophila embryo with domains of TF expression stained in various colors. The regulatory network responsible for spatial patterning along the long embryo axis will ultimately convey a unique cell fate to each of the ~100 rows of nuclei, by extracting positional information from maternally-deposited chemical cues. Curtesy of flyex.
Transcriptional regulation is noisy. Basic physical processes of TF diffusion, binding to DNA, and synthesis of molecules such as mRNA and proteins, involve small numbers of molecules and must be described probabilistically. We analyze these contributions to noise in gene expression and ask how cells manage to function reliably in the presence of such noise.
In particular, we study processes in the early development of multicellular organisms, where each cell in the organism reads out chemical cues in its vicinity and, based on them, commits to a particular cell fate – e.g. should it activate the developmental program for the wing or the nervous system? This spatial patterning of the embryo involves the cells extracting “positional information” from the chemical cues as reliably as possible given the relevant physical constraints. Our research aims to turn this intuitive picture into a precise mathematical statement.