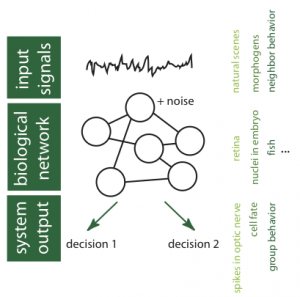
Schematic diagram of information processing in biological networks showing our main areas of study: statistics of input signals, noise and computation in network elements, network achitecture, network readout. Some biological examples shown on the right.
Many interacting systems in biology – networks of genes, signaling proteins or neurons, and even whole organisms engaging in collective behaviors – sense, transduce and process internal and environmental signals in order to guide their behavior. We are interested in the basic principles underlying this “biological computation.”
Our basic intuition is that biological computation can be analyzed with the tools of statistical physics and information theory, despite significant differences between biological and engineered information processing systems. To this end, we need to understand the statistics of input signals that the networks receive, their architecture, the elementary “computations” that they perform, the nature of intrinsic noise in the system, as well as the physical and functional constraints acting on the network.
Our group is theoretical, but most of the work is done with real data in close collaboration with our experimental colleagues. For details, select a Research subtopic from the menu.